Welcome to the 4th Phase of WGIN (2018-2023)
Defra Wheat Genetic Improvement Network:
WGIN 4 - Improving the resilience, efficiency and stability of the wheat crop through genetics and targeted trait analysis
WGIN 4 has been funded by Defra until July 2023. Due to Covid this has now been extended until July 2024!
The strategy for WGIN4 including continuing as well as new projects is shown below:
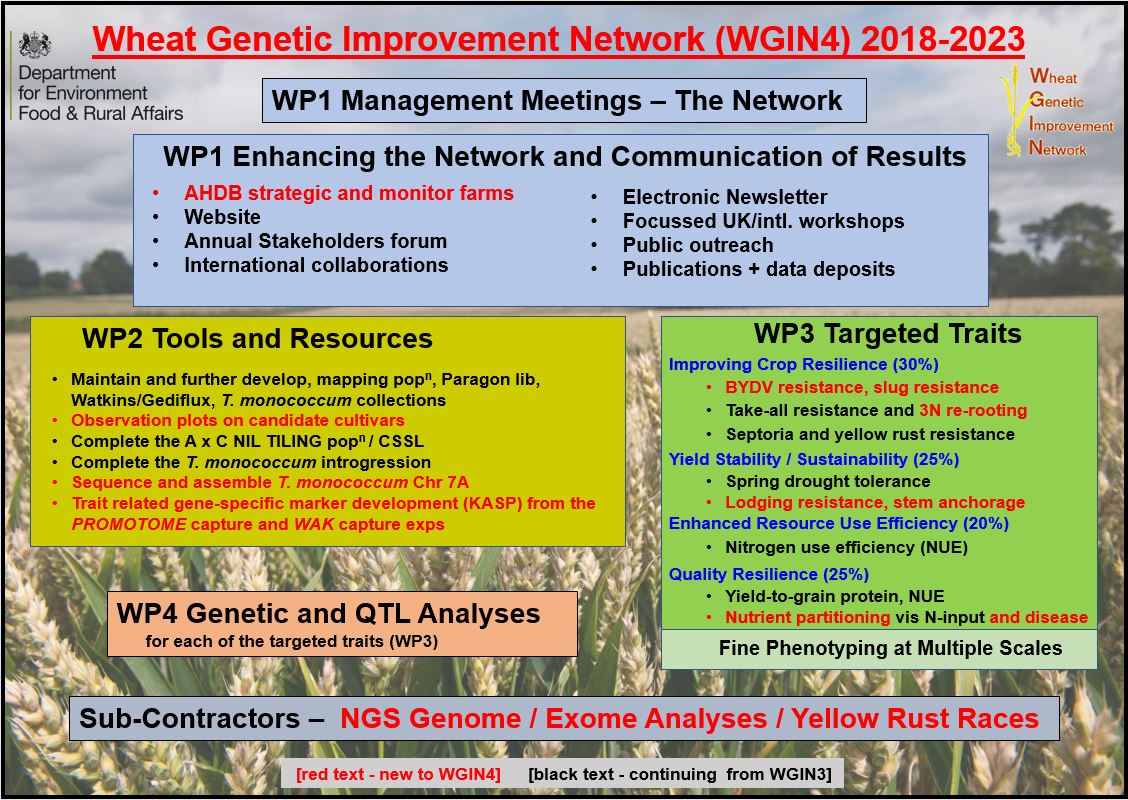
A summary of WGIN3 achievements and the WGIN4 core project can be found here.
IMPORTANT:
The 21st WGIN Stakeholders' Meeting on 8th February 2024 was another hybrid event, hosted by the JIC.
This was a particularily succesful event with great presentations, interviews with two high profile farmers, a lively panel discussion on "Climate change: the implications for breeding and the wheat crop – 10 years plus" as well as a lot of audience participation. Thanks to everyone who made this meeting into such a nice experience.
The whole meeting was recorded and can be enjoyed by clicking any of the four parts here:
part 1) "Market update" to "DSW"
part 2) "Enhancing Wheat Nutritional Quality" to "Crop growth and phenology"
part 3) "Future changes in the UK climate" to "Climate change from the farmers' perspective"
part 4) Panel discussion
PDFs of the presentations can be viewed from the Stakeholders drop-down menu by clicking on "meetings".
1) The 20th WGIN Stakeholders' Meeting on 6th February 2023 was a hybrid event, hosted by the JIC, with limited in-person attendance due to Covid. The recordings of this event can still be viewed/ downloaded from the Staekeholders drop down menu.
2) The April 2023 edition of the WGIN Stakeholders' Newsletter can be viewed here.
3) The first WGIN Wheat Yield Stability Workshop, organised as a hybrid event at the John Innes Centre and online by Clare Lister and Simon Griffiths, was held on Monday 21st March 2022. As the whole meeting was recorded it is available to watch in two parts by clicking part 1 or part 2. Enjoy!
4) The WGIN Legacy document, covering the achievements and outcomes of the first TEN years (WGIN 1 2003-2008, WGIN 2 2009-2014) is now available to peruse by clicking this link: WGIN 1&2 Legacy
5) An article by Heather Briggs about WGIN has been published in the June 2018 Edition of Arable Farming. The article can be found here.
WGIN videos:
For the first time, as part of WGIN4 we are now in the process of creating videos about various aspects of WGIN and making these available to website visitors.
As a start, you can view a 6min video about wheat crossing and the rationale for trying to introgress T.monococcum into bread wheat here.
Upcoming Events:
***
Release of compiled yield and NUE data for WGIN Diversity trial, 2004-2019
***
 The Wheat Genetic Improvement Network (WGIN) germplasm diversity trial is an example of a multi-variety, multi-N treatment series of trials conducted over multiple years. Data from the initial years (2004-2008) of these trials reported variation in yield and N-responses and contributing physiological processes (Barraclough et al. 2010; 2014). The trials have continued to the present date and have involved a large panel of modern commercial hexaploid wheats (varieties introduced between 1964 and 2016) and data has been compiled for trials from 2004-2019 and is available on this website. In most years there were four N rates, from zero to 350 kg N/ha/yr, which represents no input through to excess applied N. All trials were conducted following local commercial agronomic practice, at the Rothamsted Farm in Hertfordshire in the UK. Whilst more than 60 varieties were examined in total, a smaller subset of 15 core varieties have been grown for most years.
The Wheat Genetic Improvement Network (WGIN) germplasm diversity trial is an example of a multi-variety, multi-N treatment series of trials conducted over multiple years. Data from the initial years (2004-2008) of these trials reported variation in yield and N-responses and contributing physiological processes (Barraclough et al. 2010; 2014). The trials have continued to the present date and have involved a large panel of modern commercial hexaploid wheats (varieties introduced between 1964 and 2016) and data has been compiled for trials from 2004-2019 and is available on this website. In most years there were four N rates, from zero to 350 kg N/ha/yr, which represents no input through to excess applied N. All trials were conducted following local commercial agronomic practice, at the Rothamsted Farm in Hertfordshire in the UK. Whilst more than 60 varieties were examined in total, a smaller subset of 15 core varieties have been grown for most years.
All data can be accessed by following this link.
A forthcoming Frontiers in Plant Science paper presents an overview, and a soon to be published Defra report will give a detailed statistical evaluation.
Professor Malcolm Hawkesford (malcolm.hawkesford@rothamsted.ac.uk), Head of the Plant Sciences Department and the Designing Future Wheat Institute Strategic Programme at Rothamsted.
Honorary Professor in Plant Sciences in the School of Biosciences, University of Nottingham
***Paragon Library***
IMPORTANT:
The latest update to the Paragon Library, version 2.3, is now available for download.
WGIN has been part of an informal consortium developing NILs in the genetic background of the UK spring wheat Paragon. The collection, known as the Paragon Library, was developed at JIC and currently consists of around 350 lines. The project involves crossing different combinations of genes, QTLs and mutations into the common background of Paragon and then studying the phenotypic effects. NILs are currently available for multiple alleles of: Rht-D1, Rht-B1, Rht8, Lr19, 1BL.1RS, Yield (7B), Grain Size (5A, 7A) and more than 10 Heading Date QTL. Further lines will be added in the future.
The Paragon Library lines have been genotyped on the Axiom 35K Breeders' Array and data for about 300 lines is now available to download here and the figure to the right indicates the format of the data. Please note that this is a substantial update, version 2.3 . More details can be found under the 'Resources' tab and the 'Paragon Library' submenu.
Seed collected from the genotyped plants are now available for distribution on request and we hope they will eventually become available through the Germplasm Resources Unit (GRU) at JIC.

.jpg)
